Pitt researchers map Huntington’s protein
(neurowiki2012)
Pitt researchers are closer to curing a nerve cell disease after creating a new protein model.
For the first time, a team of 10 Pitt researchers from the School of Medicine mapped the tiny fibers of the huntingtin protein, a single mutated protein on the fourth chromosome that causes Huntington’s disease, an incurable hereditary disease. The researchers found that the “huntingtin exon 1” fibers have interlocking strands of polyglutamine, another type of protein, which clump together and lead to the death of neurons.
Their findings, which were published in the Feb. 9 weekly issue of Proceedings of the National Academy of Sciences, will help develop future treatments for the disease. According to Jennifer Boatz, a graduate researcher and one of the authors of the study, the huntingtin protein structure will show researchers how the disease progresses.
“We know that huntingtin fiber formation is linked to Huntington’s disease progression, so if we can figure out how these fibers form, we can better understand how Huntington’s disease progresses in patients,” Boatz said in an email.
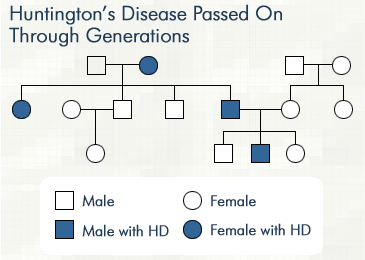
Most people with Huntington’s disease develop symptoms in their 30s and 40s, though an onset of symptoms can begin earlier. According to Medical News Today, about one in every 10,000 Americans has Huntington’s disease, and more than 200,000 people in the United States are at risk for inheriting the disease. The disease deteriorates nerve cells in the brain, causing motor and cognitive deficits.
“There is no cure for Huntington’s disease, therefore, symptoms that patients are having are targeted and treated,” Tammy Makoul, a social worker for the western Pennsylvania chapter of the Huntington’s Disease Society of America, said in an email.
For the study, the research team made versions of the huntingtin protein based on the faulty version that causes Huntington’s and studied how the proteins behaved. In the lab, the proteins clumped together to form fibrils, as they do in the brain of someone who has Huntington’s Disease, Patrick Van der Wel, the study’s senior investigator, said.
The team then placed the samples of their protein into an NMR — or nuclear magnetic resonance — instrument to probe the structure of atoms inside the protein aggregates. With the sample spinning inside the NMR up to a million rotations per minute, the NMR shows researchers the proximity of different atoms within the protein aggregates, Van der Wel said.
“So you know that atom one is close to atom five but not close to atom two. Then you collect that for a bunch of different things and you start to build a picture,” Van der Wel said.
Proteins, which are sequences of organic compounds, fold into particular structures that have specific functions. A stretch of a particular amino acid called glutamine or “Q” is approximately 20 glutamine amino acids long in some proteins but is 40 or more “Q” amino acids long in Huntington’s disease patients, according to Van der Wel.
The “Q” amino acids overlapped in the NMR readings, Van der Wel said, making it difficult for scientists to study the aggregate structure.
“Nature made it in some way that it’s hard to study, so we had to develop some new ways of dealing with this that work around this limitation of the system,” Van der Wel said.
Pitt researchers checked their new detailed structure of the protein aggregates against previously proposed models which Van der Wel said were based mostly on speculation.
The research team concluded that none of them fit their data. With their results, the team built an improved version of past models and plans to eventually conduct more experiments to refine their proposed model.
According to Boatz, scientists can potentially design structure-based drugs to prevent the mutated huntingtin fibers from forming if they know the exact mechanism of how the fibers form.
“The structural information that we have gathered on huntingtin exon 1 so far is just one of many first steps in the direction of treating Huntington’s disease,” Boatz said.
The expansion of the “Q” amino acid sequence appears in other neurodegenerative disorders, causing problems in motor function, such as involuntary jerking movements, impaired gait and difficulty with speech. Because of this commonality, Van der Wel hopes the research team’s work on Huntington’s disease can lead to better understanding other similar diseases.
“We want to understand how this happens in part to help understand Huntington’s disease and in part to understand in general, ‘how does this happen and why, and how do proteins do this?’” Van der Wel said.
For Makoul, the research team’s improved model offers hope for potential drugs and other treatments for a disease that was once deemed incurable. So far, the work researchers have done on Huntington’s is advancing medicine and providing families of Huntington’s patients with hope, Makoul said.
“Researchers are the ones that those with HD, and their families are looking toward to find a cure,” Makoul said. “Any research movement forward in a neurodegenerative disease will be helpful with others.”
Recent Posts
SGB addresses concerns about ICE presence on campus, hears SJP lawsuit against administration, approves governing code bill
At its weekly meeting on Tuesday at Nordy’s Place, Student Government Board heard concerns about…
ACLU of Pennsylvania sues Pitt over SJP suspension
The ACLU of Pennsylvania filed a federal civil lawsuit against the University of Pittsburgh and…
Marquan Pope: The ultimate shark
One of the most remarkable things about sharks is that an injury doesn’t deter them.…
Who Asked? // Do we really get a summer vacation?
This installment of Who Asked? by staff writer Brynn Murawski mourns the seemingly impossible perfect…
Notes From an Average Girl // Notes from my junior year
In this edition of Notes From an Average Girl, senior staff writer Madeline Milchman reflects…
Meaning at the Movies // The Power of the Movie Theater
In this edition of “Meaning at the Movies,” staff writer Lauren Deaton discusses her love…

